Günter Seyfried
Clemens Grabher
Dominik Bartenstein
2004
Multi-step Polycinease Film Transformation (PFT)
(binary conversion, oligo design, gene synthesis, E. coli transformation)
Here, we describe PFT as a method for the conversion of binary data into DNA sequences and subsequent storage of artificial information in a living organism. Our in-house software converted animated gifs into a DNA sequence that was transformed into a laboratory bacterial strain of E. coli bacteria, relying on oligodeoxyriboncleotide assembly, DNA polymerase chain reaction (PCR) and electroporation. The synthetic gene, assembled from 28 oligonucleotides, was PCR amplified and cloned in a bacterial DNA vector containing an ampilicin-resistance gene, the pUC origin of replication and the red fluorescent protein (RFP) as a marker. After ligation and transformation and propagation in Escheria coli, recovery of the introduced DNA vector yielded the correct plasmid containing the originally designed information.
Experimental:
In the first step, we converted the animated gif ‘polycinease_testfilm_05.gif’ with our in-house software, the dnaspitter.py v 1.0b, into a string of gatc. From the output we were able to design the oligos using the online version of the DNAWorks software. The oligos were annealed, the assembled DNA sequence was amplified by PCR and the resulting DNA fragment was cloned into a bacterial vector, which contained the ampilicin-resistance gene, the pUC origin of replication and the RFP to enable visual analysis. This was followed by the transformation of this vector into E. coli; using the electroporation method, that utilizes an electric pulse to permeabilizethe bacterial cell membrane, allowing the DNA to enter the cell. Thus, the converted film sequence floated as part of the circular DNA vector molecule inside the E. coli cells (1).
Transformed bacteria were grown on LB agar plates, containing ampicillin to maintain selective pressure on bacteria that had taken up the plasmid DNA. 12 hours of incubation at 37°C allowed the bacterial cells to grow for approximately 36 generations. Analysis of single bacterial colonies by UV-microscopy revealed that most colonies had integrated our DNA vector resulting in expression of the RFP as a visual marker for successful transformation. One single colony was inoculated in bacterial growth medium and grown overnight at 37°C. Plasmid DNA extracted from this overnight culture was isolated by alkaline lysis and ethanol precipitation. By enzymatic restriction of the plasmid we could confirm the presence of the initially assembled DNA sequence of 432 base pairs representing the animated gif. Furthermore, DNA sequencing showed that the isolated sequence was identical with the sequence we transformed into the E. coli earlier.
Conclusion:
We performed this pilot experiment to show that basic artificial information can be converted into DNA sequences. In the form of DNA the information can be stored in bacterial cells that are unable to recombine DNA. The lack of recombination in the bacterial cells allowed the recovery of the original information. (Organisms with the potential to recombine DNA will introduce changes in the DNA sequence, a mechanism to maintain genome plasticity and thus adaptation to changes of the environment.)
(1) Willem P.C. Stemmer, Andreas Crameri, Kim D. HA, Thomas M. Brennam and Herbert L. Heyneker: Single-step assembly of a gene and entire plasmid from large numbers of oligodeoxyribonucleotides. Gene, 164 USA (1995) 49-53.
David M. Hoover, Jacek Lubkowski: DNAWorks: an automated method for designing oligonucleotides for PCR-based gene synthesis. Nucleic Acids Research, 2002, Vol. 30, No. 10e43. Sambrook, J., Fritsch, E.F. and Maniatis, T. (1989) Molecular Cloning, A Laboratory Manual. Cold Spring Habor Laboratory Press.
polycinease_testfilm_05.gif ![]()

polycinease_testfilm_05.gif (zoomed)

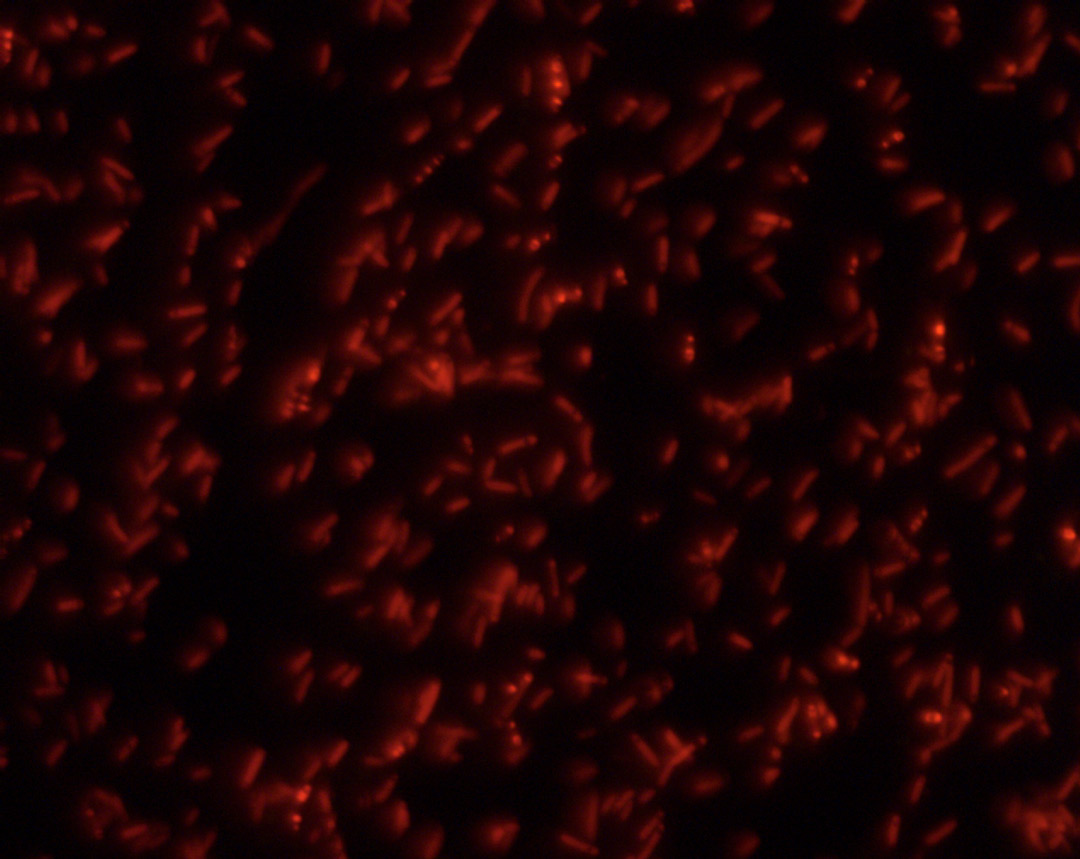
The bacteria successfully picked up our vektor, as the rfp indicates